This article first appeared on the Medium website but has been moved to this blog… There’s also now an Inferred Segments Generator tool
Chromosome mapping is the process of using the data from matches to assign segments of your chromosomes to specific ancestors — from grandparents to more distant forebears. DNA Painter is a simple web-based tool for chromosome mapping that I built and launched this summer.
Inferred matching is a simple technique that can help you use sibling or parent matches to assign additional segments to ancestors.
While all inferred segments should be considered provisional — and I mention several hazards below — you may find this technique helps accelerate the process of identifying your segments by grandparent. What follows is a short step-by-step explanation of how I’ve used it for my own DNA.
My father had a German Jewish father and an English mother. Due to current patterns in the popularity of autosomal DNA testing, I wasn’t surprised to find that the vast majority of my father’s matches were from his Jewish father’s side. Indeed, due to endogamy, it can be quite hard to find cousin matches from his English side in amongst the others.
I am in the fortunate position of having had both parents tested, but this technique can also be used with siblings.
I realised quite early on that I can use my parents’ matches to infer which grandparent I myself inherited these DNA segments from.
While it can be confusing, the essence of inferred matching is very simple:
- I inherited one half of my DNA from my father. This DNA was made up of a random selection of segments from his maternal and paternal chromosomes.
- So if my father shares a segment (e.g. from position 111,111,111 to 131,111,111 on chromosome 1) with a paternal cousin, and I don’t share this same segment with this cousin, then I can infer that I inherited this segment from his mother’s side, not his father’s.
First example: my father’s 2nd cousin
Theresa* is my father’s 2nd cousin. She’s the granddaughter of his great-aunt, and she and my father share about 300cM across 13 segments. I share about 80cM across 4 segments with Theresa, but the other 220cM are precious too!
- I know that the segments I share with Theresa came to me via my great-grandfather (the common ancestors being his parents)
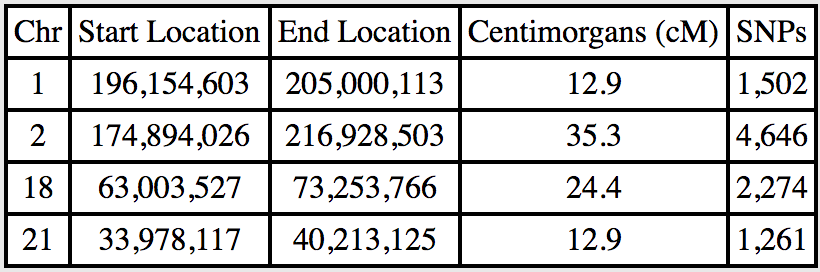
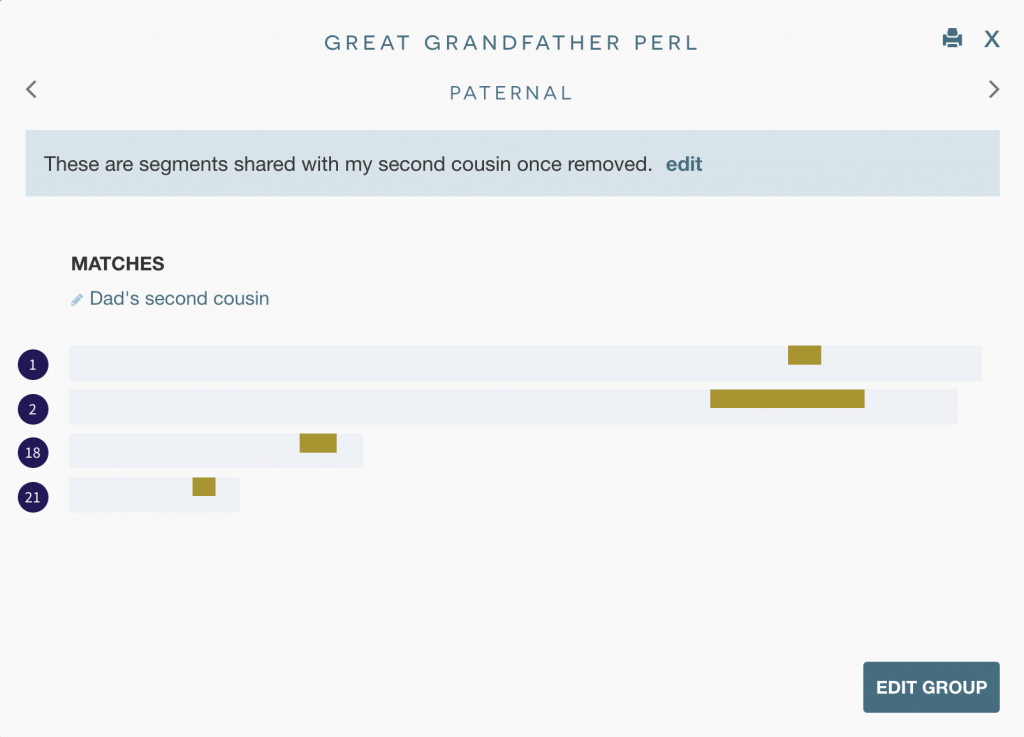
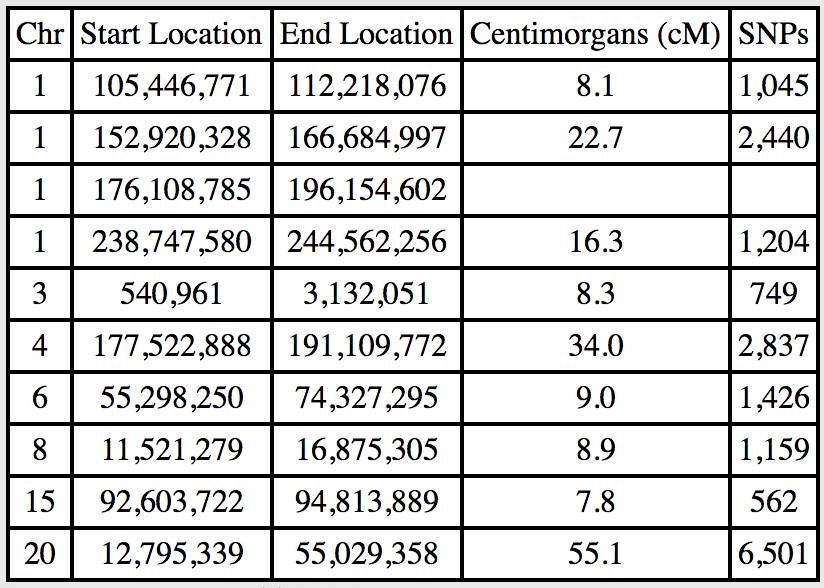
- For the segments I don’t share with the cousin, I can use inferred matching. Here are the segments my dad shares with her:
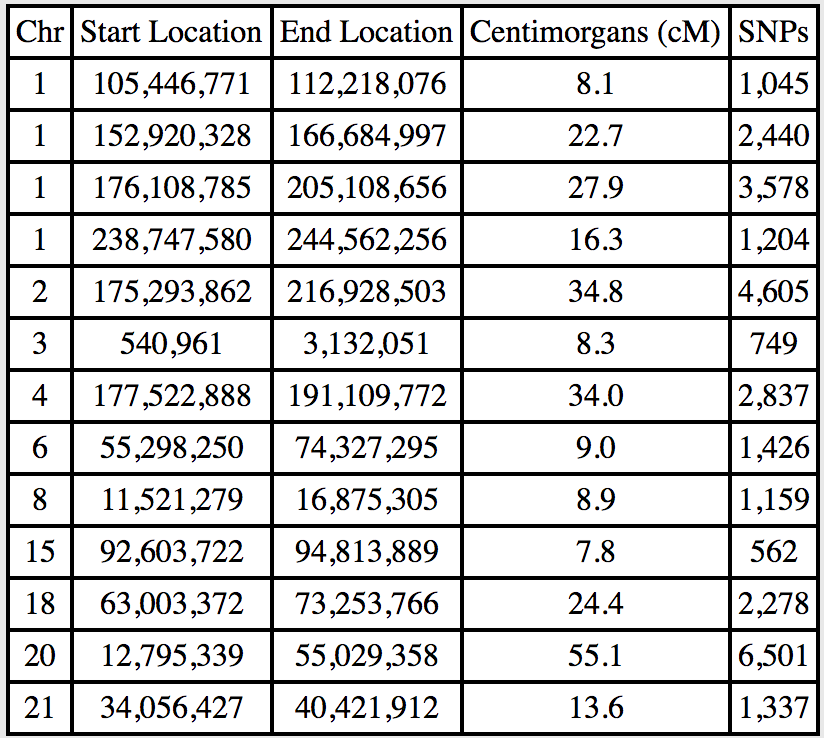
That’s a big difference: 13 segments vs. just four that I share with her! While a handful of these segments are under 10cM, there are some huge ones, including a 55.1cM segment on chromosome 20.
In order to use this data in my own DNA profile, I first copy it and leave only the segments that I don’t share with the cousin. Note that on Chromosome 1 there’s a segment that I share, but only the last part. I don’t have an easy way to calculate the cM or SNP values for this partial segment so I leave that bit blank:
I know that the chromosomes I inherited from my father were made up of a random combination of segments from his mother and father. Since I’ve established I don’t share the segments that he got from his father, I can infer quite clearly that I inherited these segments from his mother, my grandmother.

This gives me an additional 220cM that I can assign to a specific ancestor (albeit a very close one, my grandmother).
Second example: my father’s half brother
David and my father share 1,826 cM across 34 segments. I can use the same simple process to expand significantly the number of segments I can paint on my own profile:
- Any segments I share with David came from my grandfather (this amounts to 1,101cM)
- There are segments my father shares with David, but which I don’t. Since David and my father share these segments on their paternal chromosomes, I must have inherited them from my father’s maternal side. I can therefore infer that they came from my grandmother
Third example: unknown matches
My father has has a fair number of untraced matches. While these are relatively small (under 100cM shared in total), there are a good number of matches with long segments (40cM+) who match with both my father and his half-brother, but not with me.
Because of how I’m related to my half-uncle, I can narrow down these matches to my paternal grandfather. Due to the separation of my genetic mix (my paternal grandmother was English with no known Jewish ancestry, while my paternal grandfather was German Jewish), I can be more sure of this than most people.
So even though I don’t know how we’re related to these untraced matches, I can take any segments my father matches with them and add these to the list of segments that I can infer that I inherited from my grandmother (since I don’t share them with the untraced matches or my father’s half-brother).
I can follow the same process, substituting my half-uncle with every other close match where I can narrow down my relationship with them to a grandparent, such as:
- my mother’s paternal cousin (I know the segments we have in common came from my maternal grandfather)
- my second cousin, the daughter of my mother’s maternal first cousin (I know the segments we share came via my maternal grandmother).
Using siblings
Differences in sibling matches can be used in exactly the same way. To put the first two examples together: Theresa is my father’s second cousin via his paternal grandfather. My father and his brother David match Theresa on different segments. For those segments on which David matches Theresa, but my father does not, I can infer that my father inherited these segments from his paternal grandmother since David got them from his paternal grandfather!
Possible hazards and pitfalls
There are a few potential issues you might want to look out for (and I’m sure there are others I haven’t thought of yet):
- You might be related to the match in two or more ways, in which case you can’t say ‘I share these segments with the match; therefore they came from this ancestor’
- Thresholds might come into play. It’s possible that you might share valid small segments with a match that don’t show up in the default search, and so you might infer incorrectly that these segments came from somewhere else. In my experience these errors tend to be uncovered quite quickly when a match comes in to contradict them.
- Within DNA Painter, I always adjust the certainty level on inferred matches to ‘possible’ or lower since they are inherently speculative (as far as I know, it’s not possible to triangulate an inferred match!)
I haven’t yet completed this process, but even with just a handful of matches, I’ve been able to map a good number of segments to my grandmother as illustrated here:
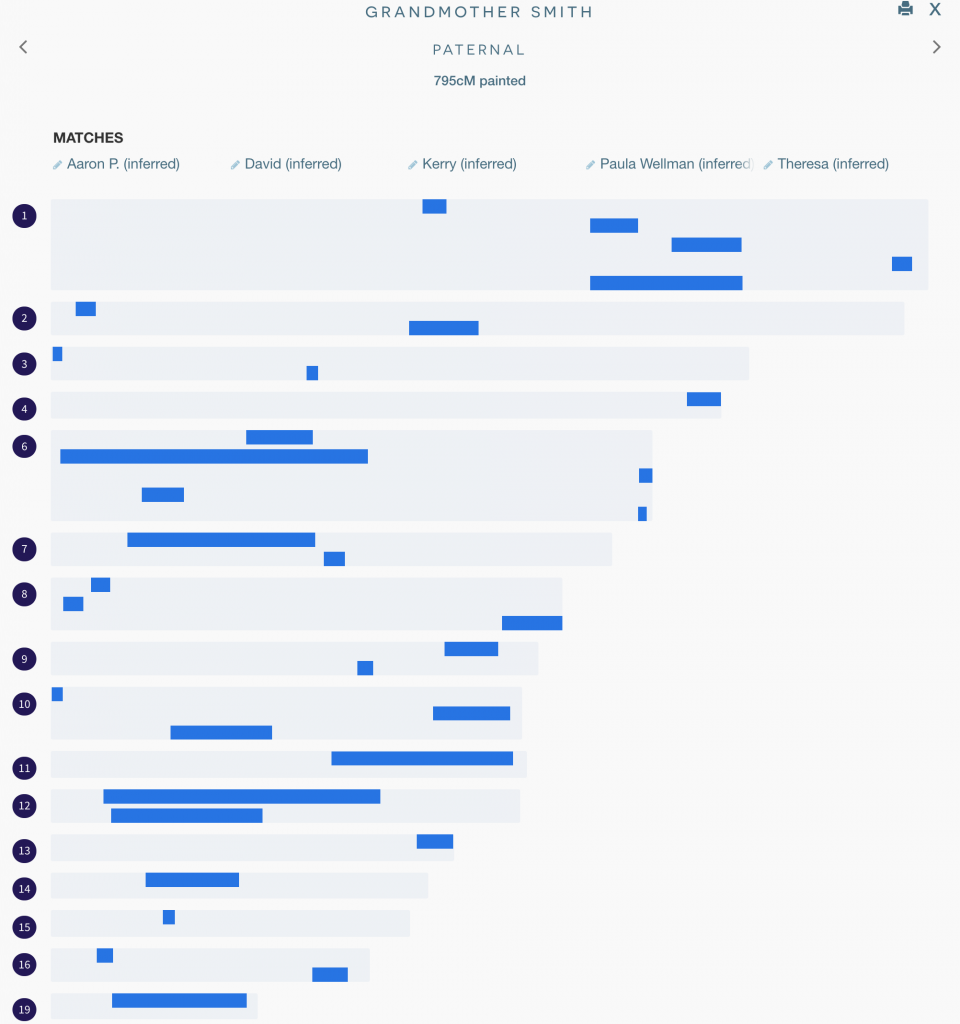
I hope this has been helpful; all feedback welcome!
Jonny
* all names have been changed
Some further reading: Louis Kessler has an article on how his Double Match Triangulator now accommodates inferred matching.
Some more further reading: I have a follow-up article: More tips for inferred chromosome mapping
Contact info: @dnapainter.bsky.social / jonny@dnapainter.com