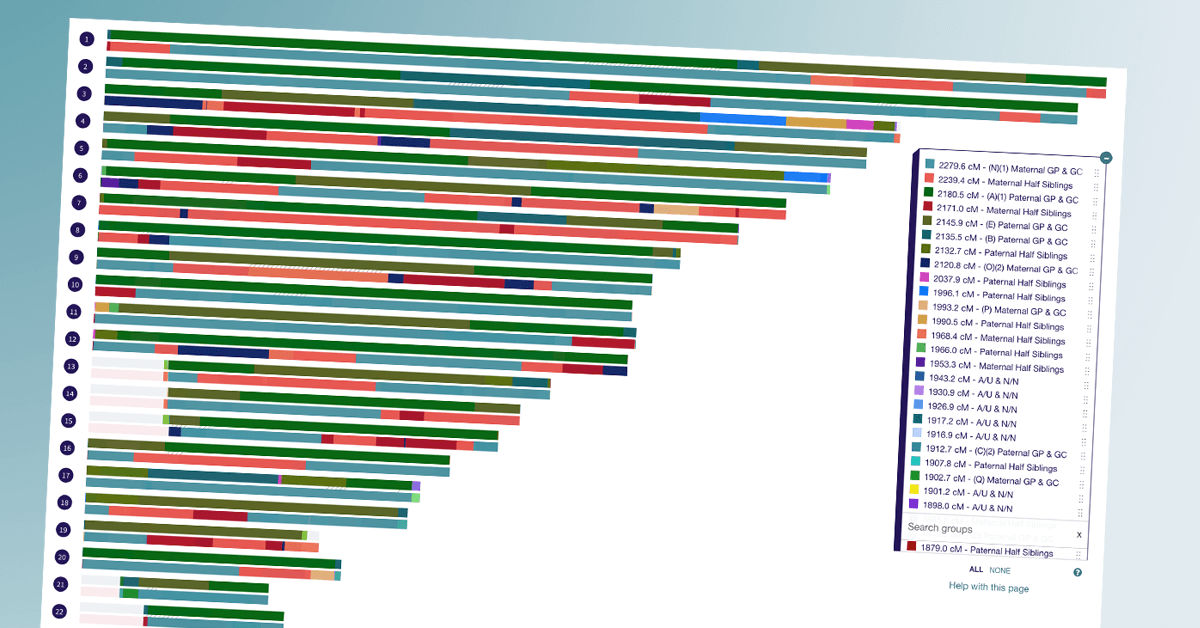
A few weeks ago, I got an email from a DNA Painter user that piqued my interest. Cody Ely had put together an anonymised chromosome map made up of a collection of matches where the genealogical relationship between the tester and the match is known.
Cody thought other users might find it useful to see the DNA shared due to different relationships visualised in a map, and I agreed. You can find the Library of Matches listed under Tools at DNA Painter. Below are some answers to questions I had for Cody.
Q&A
Please introduce yourself
Hi! My name is Cody Ely, and I am a genetic genealogist. I have been doing genetic genealogy for about eight years now. In addition to the work I have done over the years on my own family, I have also been able to use my experience and skill set to help others with their traditional or genetic genealogy needs. I am also a member of ISOGG.
How and when did you first become interested in genealogy and DNA?
Like a lot of people, I first became interested in genealogy and DNA for ethnicity reasons. I got tested at 23andMe, which did answer some ethnicity questions I had. However, I quickly realized that my DNA matches were the most fascinating part of having been tested. The idea of being able to figure out not what, but who I am made of, was one of the most incredible things I had ever come across. I still think so! It really all seems like science fiction, but it is really science fact!
How did you get the idea to create the Library of Matches?
I got the idea to create this a few years ago when I was working on an unknown grandparent case. I was trying to determine which of two brothers was the real grandfather, and had a match that was a grandchild of one of the brothers. I knew this match would either be a half-first cousin, or a second cousin to the test taker. At 333.8 cM, it was not definitive based on cM alone.
Having mapped out segments for years at this point, I had seen what the segment patterns tended to look like for numerous second cousin matches. However, I had never seen what a real half-first cousin looked like mapped out. I was curious if there would be any differentiating clues by seeing the segments of this relationship type that might help me be able to better predict if this match was a half first cousin, or a second cousin. The idea of having a visual reference guide of real shared segments for as many relationship types as possible just snowballed from there. To the best of my knowledge, the Library of Matches is unique in that it is the first relationship prediction tool that allows the user to actually see the shared segments in the data set.
Could you talk through a couple of examples of how you think the Library of Matches could be used?
For people of any experience level, the Library of Matches is first and foremost a relationship prediction tool like the Shared cM Tool. Rather than entering the cM amount, here you will just scroll down the key on the right and find the example match that is closest in cM to your unknown match. That will quickly give you an idea of the possible relation. I also recommend looking at the other matches in the same cM range for additional possibilities. You could then display the matches you are interested in on the map, one at a time, and see if the segment pattern has any similarities to your unknown match.
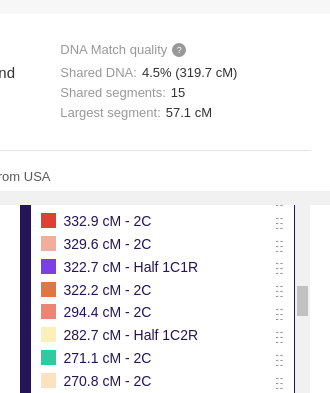
In addition, the Library of Matches could also be used to learn more about different relationships by seeing what the shared segments tend to look like for each relationship type. If you are in need of seeing real examples of a specific relationship (or just curious about certain ones), you can type that relationship’s abbreviation into the ‘Search groups’ field on the key. This will quickly show you what examples the Library has for the relationship you are interested in.
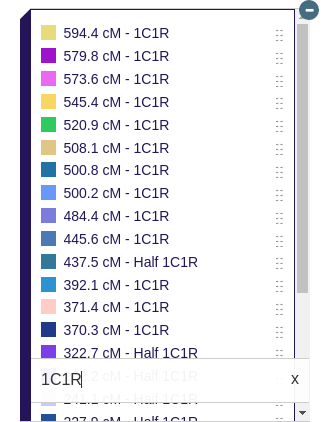
There is also another use of the Library that I am very excited about. You can display different matches on the map at the same time to see phased genomes, full siblings, and unique relationships. For phased genomes, I have made a classification system to assist the user in knowing which matches should be displayed at the same time.
Each paternal grandparent match in the key with the same letter in parenthesis at the beginning of the name can be displayed on the map at the same time to visualize a real phased paternal genome. This is great for users that want to see example male recombination patterns. Likewise, maternal grandparent matches with the same letter at the beginning can be displayed at the same time to visualize real female recombination patterns.
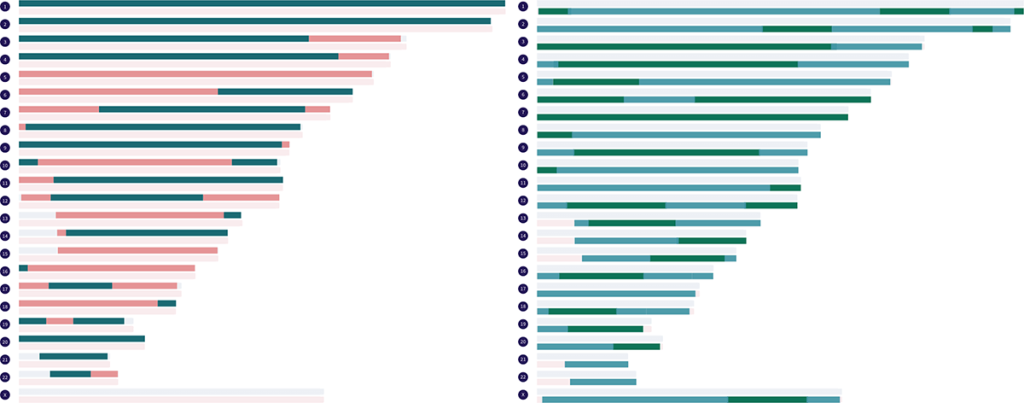
Paternal and maternal grandparent matches can also be displayed on the map at the same time to simulate fully-phased individual genomes. The two paternal matches you choose to display must have their own same beginning letter in parenthesis, and the two maternal ones you choose must have their own same beginning letter. There are currently 25 fully-phased genomes that can be simulated from the real paternal and maternal grandparent matches in the Library.
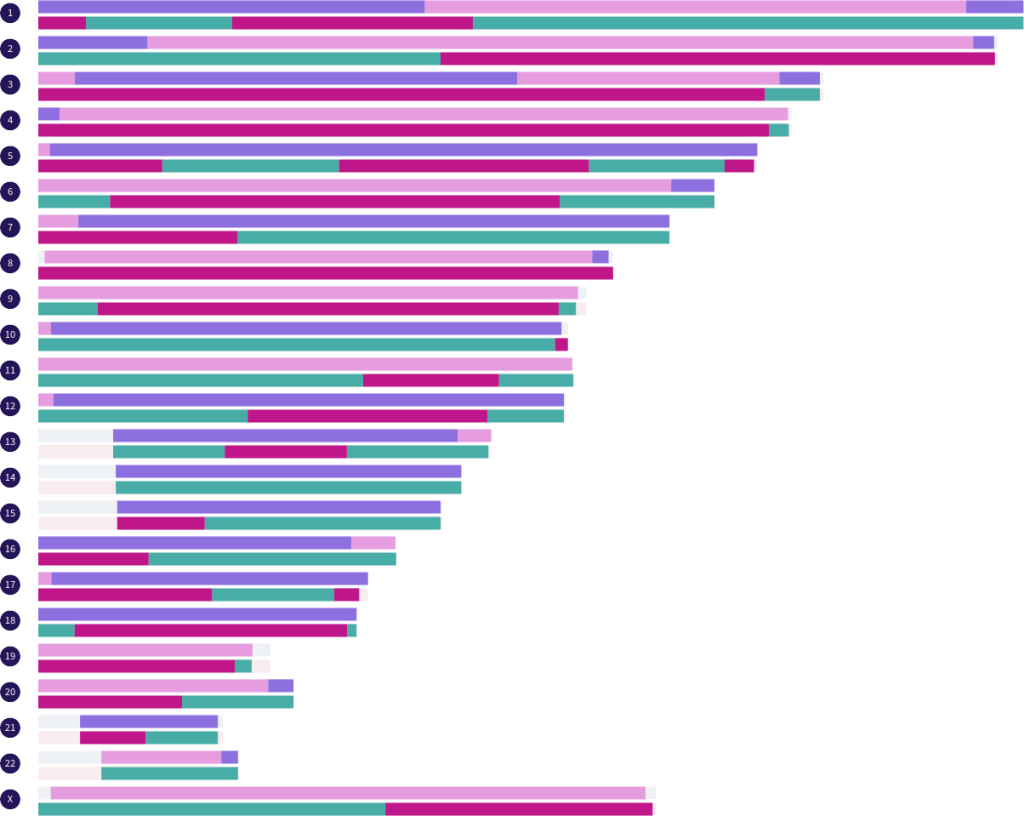
For those who are interested in seeing simulated examples of full sibling matches and the fully-identical regions they share, you can display a single paternal half siblings match and a single maternal half siblings match of your choice on the map at the same time. There are currently 121 full sibling matches that can be simulated from the real half sibling matches in the Library.

At the bottom of the key, under the divider line, is a real full siblings match that I have phased by grandparents. The paternal side segments and the maternal side segments are in their own groups. Both of these groups can be displayed on the map at the same time. Hovering over a segment on the map will show you which grandparent the segment came from.
Unique relationships, such as second cousins on both sides, can be simulated by displaying a single paternal side second cousins match and a single maternal side second cousins match of your choice on the map at the same time. There are currently 176 second cousins on both sides matches that can be simulated from the real second cousin matches in the Library. Similarly, other unique relationships can be simulated by displaying the appropriate matches at the same time.
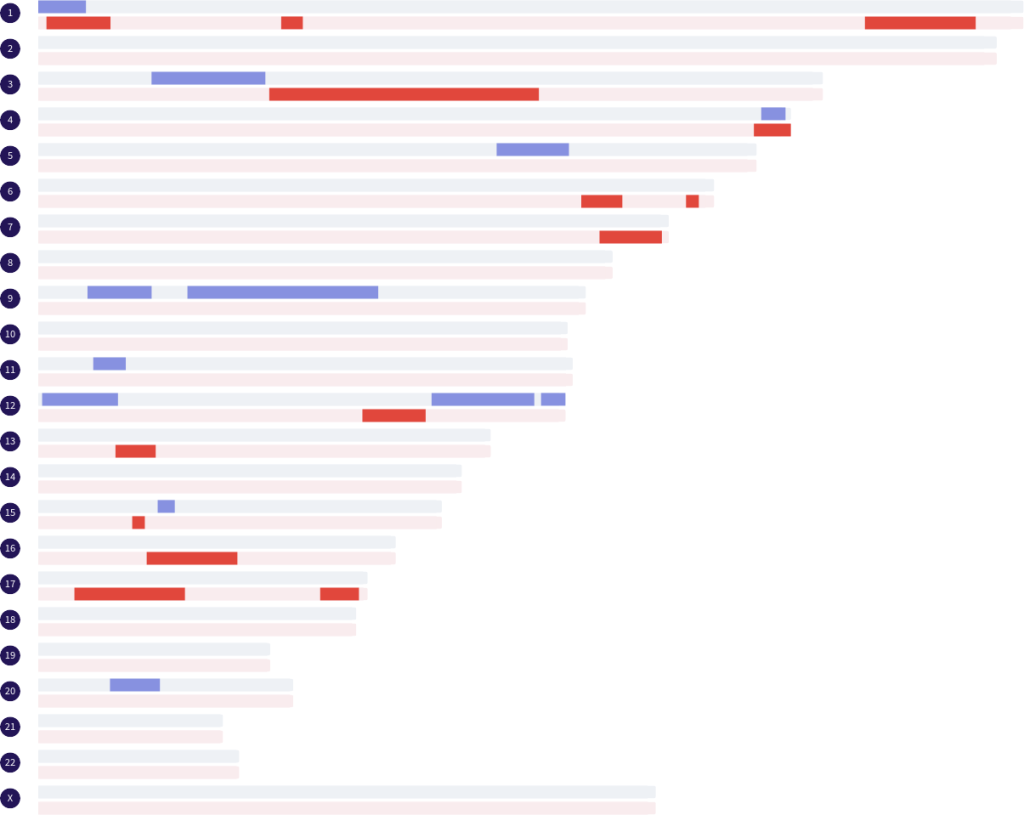
You also mentioned the term ‘segment patterns.’ Can you explain more about what you mean by this?
Certainly. When using the term ‘segment patterns,’ I am referring to the number of segments that are shared, the various sizes (in base pairs) of the segments, and how many segments there are on a chromosome. That is a written explanation of something that can be rather easily determined by looking at the segments mapped out on a chromosome map or chromosome browser. I do not count any segments on the X chromosome because the inheritance pathway is not common to all matches.
Users of the Library will notice that I have also specifically labeled grandparent and half sibling matches as paternal or maternal. As a result of my work in creating the Library, I have found that you may be able to predict if a grandparent or half sibling match is paternal or maternal, and also distinguish between grandparent and aunt/uncle matches depending on the segment pattern. This could be very helpful for adoptees.
I would like to note that Andrew Millard, Kitty Cooper, and the Williams Lab at Cornell University have also each done work supporting differentiating segment patterns among these specific relationships. I believe my findings below are consistent with their findings, which furthers my belief that segment patterns can be useful as an additional relationship prediction method. All of my findings are derived from the Library of Matches data set. In addition, I am currently working on segment pattern calculations for all of the relationship types represented in the Library of Matches.

Paternal Grandparents
- Segment appearance: tendency toward long segments with usually no more than two segments per chromosome
- Number of autosomal segments shared: 19-27 (average 23)
Maternal Grandparents or Paternal Half Siblings
- Segment appearance: tendency toward long and medium segments with some small segments
- Number of autosomal segments shared: 22-43 (average 32)
Maternal Half Siblings or Aunts/Uncles of either side
- Segment appearance: tendency toward numerous small and medium segments with some long segments, and multiple segments per chromosome
- Number of autosomal segments shared: 35-56 (average 47)
While there is some overlap in the data between the groups above, there is a clear difference between paternal grandparents and maternal half siblings or aunts/uncles of either side. Below is a quick example of how we can use the concept of segment patterns to predict a match:
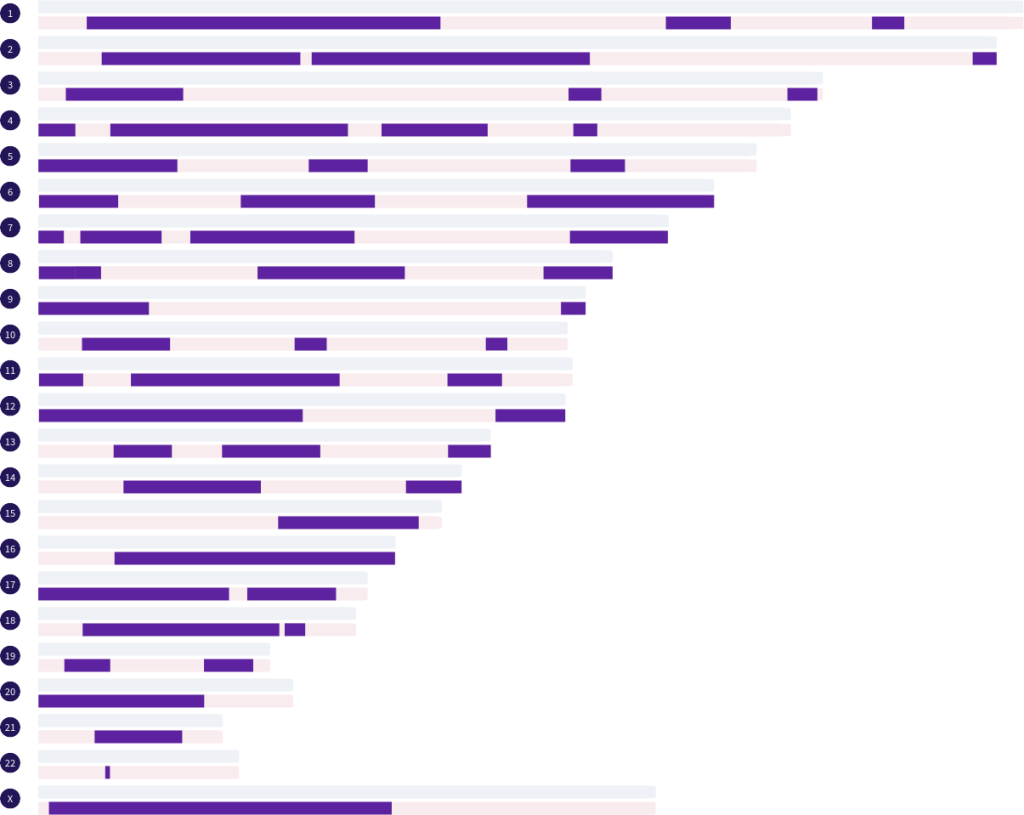
In the image above we have a match that shares 1953.3 cM. Based on cM, we can feel confident that this is either a grandparent match, an aunt/uncle match, or a half sibling match. But, which one? By analyzing the segment pattern, we can quickly rule out some possibilities. There is likely no way this match could be a grandparent. There are simply too many segments. What we see here is a total of 53 autosomal segments, with numerous small and medium segments, and multiple segments per chromosome. According to the Library of Matches data set, this pattern suggests this match is either a maternal half sibling or an aunt/uncle of either side. In reality, this match is a maternal half sibling.
The concept of segment patterns can be used on matches from all testing companies. While AncestryDNA does not allow test takers access to the shared segments, they do report the number of segments, which can be compared to the findings above. I welcome any feedback on using segment patterns to predict relationships, and on the Library of Matches in general. I would love to know if other people find it to be helpful and in alignment with their own matches.
How can people get in touch with you if they’d like to?
Feel free to message me through my Facebook page at facebook.com/codyelygenealogy or email me at ydoc14@yahoo.com
Contact info: @dnapainter.bsky.social / jonny@dnapainter.com